CLONE 082514 humKappaCasein(Kex+)
Revision as of 07:40, 5 September 2014 by Rachel Linzer (talk | contribs) (→Experiment 082514_humKappaCasein(Kex+) Cloning)
Experiment 082514_humKappaCasein(Kex+) Cloning
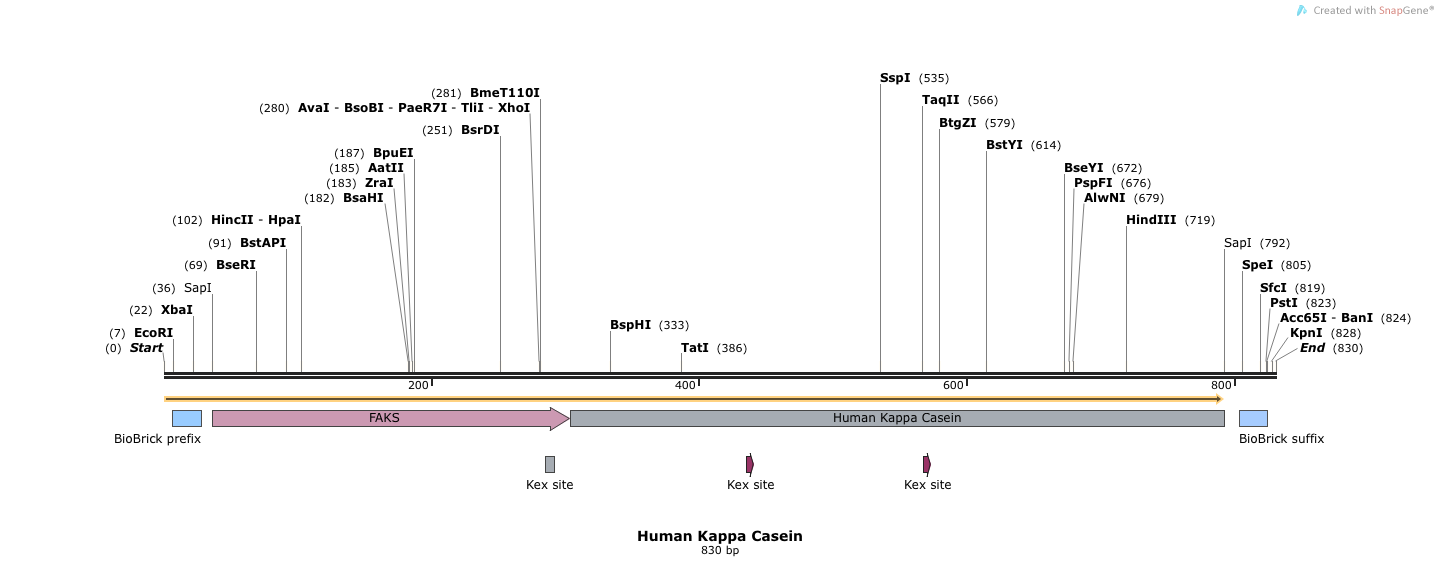
- Human Kappa Casein (BB.SapI.humKappa(Kex+).SapI.BB)
5'- TACACGGAATTCGCGGCCGCTTCTAGAGGCTCTTCTATGAGATTCCCATCTATTTTCACCGCTGTCTTGTTCGCTGCCTCCTCTGCATTGGCTGCCCCTGTTAAC ACTACCACTGAAGACGAGACTGCTCAAATTCCAGCTGAAGCAGTTATCGGTTACTCTGACCTTGAGGGTGATTTCGACGTCGCTGTTTTGCCTTTCTCTAACTCC ACTAACAACGGTTTGTTGTTCATTAACACCACTATCGCTTCCATTGCTGCTAAGGAAGAGGGTGTCTCTCTCGAGAAAAGAGAGGCCGAAGCTGAAGTCCAAAAC CAAAAGCAACCAGCTTGTCATGAAAACGACGAAAGACCATTCTACCAAAAGACTGCCCCATACGTTCCAATGTACTACGTTCCAAACTCTTACCCATACTACGGT ACTAACTTGTACCAAAGAAGACCTGCTATCGCCATTAACAACCCATACGTCCCAAGAACTTACTACGCTAATCCAGCTGTTGTTCGTCCACACGCTCAAATTCCA CAAAGACAATATTTGCCTAACTCTCACCCACCAACCGTTGTCAGAAGACCAAACTTGCATCCTTCTTTCATCGCTATCCCACCAAAAAAGATCCAAGACAAGATT ATCATCCCAACTATCAACACTATCGCCACCGTTGAACCAACCCCAGCTCCTGCCACCGAACCAACTGTCGATTCTGTTGTTACTCCAGAAGCTTTCTCCGAATCT ATCATTACTTCTACTCCAGAAACTACCACTGTCGCCGTCACCCCACCAACTGCTTAGGGTAGAAGAGCTACTAGTAGCGGCCGCTGCAGGTACCA - 3'
- This sequence has 5' and 3' SapI sequences, followed by full length FAKS, followed by human Kappa Casein (Kex+). Electra cloning cleaves at SapI sites and ligates simulatenously.
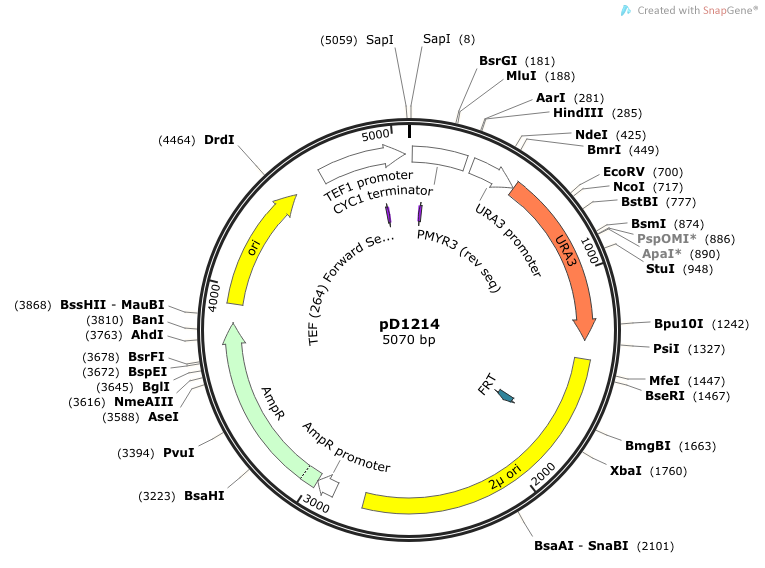
- We cloned into pD1214. This vector was ordered from DNA2.0 and is linearized upon arrival. Our insert is treated with Electra enzyme mix which cuts and ligates our "SapI.gene.SapI" and inserts it into the pD1214 vector.
- SapI treated DNA has an ATG overhang at the 5' end and a GGT overhang at the 3' end. Our complete vector will have the alphafactorfull signal sequence, which leads into another Methionine (M), followed by the rest of the human kappa casein protein.
All you need to know about the Electra system.
August 25, 2014
- Successfully cloned SapI.hKcasein.SapI into the electra daughter vector (pD1214).
Reaction: 1uL pD1214:FAKS (20ng) 1uL (20ng) of BB.SapI.humKappa(Kex+).SapI.BB 2uL Electra Buffer 1uL of Electra enzyme mix 15uL of Water ---- 20uL, Room Temperature, 20min.
- We stored the cloning reaction at -20 degrees C and waited to transform E. coli cells until August 26, 2014 (Nikola and Johan performed this) since we did not have competent cells available.
August 28, 2014: Midiprep of humKappaCasein(Kex+) in pD1214
- Location: BioCurious
- Aims: Extract the plasmid DNA from O/N culture of E. coli for further characterization of and experimentation with humKappaCasein(Kex+) in pD1214.
- Materials and Methods: Followed protocol for Zyppy Plasmid Midiprep Kit, Zymo Research: Media:D4025i.pdf.
September 1, 2014: DNA quantification of humKappaCasein(Kex+) in pD1214
- Participants: Advait, Patrik, Johan, Minakishi, Rachel, Lafia, Mac
- Location: BioCurious
- Aims: Quantify and check purity ratio of humKappaCasein(Kex+) in pD1214 (midiprepped as above, August 28, 2014) in preparation for submitting to Sequetech for sequencing.
- Materials & Methods: All measurements taken on BioCurious Beckman Coulter DU 640 spectrophotometer, DNA/Oligo setting, using 100ul cuvette.
Measuring plasmid DNA diluted 1:100: Blank = 1ul Zyppy kit elution buffer + 99ul diH2O Sample = 1ul plasmid DNA + 99ul diH2O
- Results: Spectrophotometer gave DNA concentration readings of 0. Repeating with a lower dilution factor.
Measuring plasmid DNA diluted 1:10: Blank = 10ul Zyppy kit elution buffer + 90ul diH2O Sample = 10ul plasmid DNA + 90ul diH2O
- Results:
- Read 1: 42.158 ug/mL; 260/280 ratio = 1.9291
- Read 2: 42.433 ug/mL; 260/280 ratio = 1.9382
- Conclusions:
- DNA concentration looks good; 260/280 ratio a bit high ("pure" DNA = 1.8, "pure" RNA = 2.0).
- Construct ready to send to Sequetech for sequencing.
September 4, 2014: Setting up 05Sep14 DNA pick-up by Sequetech to sequence humKappaCasein(Kex+) in pD1214
- Location: BioCurious
- Aims: Confirm correct DNA sequence of our alpha factor - human kappa casein insert into pD1214 (as mapped, above.)
- Sequencing Strategy & Methods:
- Sequencing sample hkckplus, an aliquot of August 28, 2014 midiprep, to be sequenced in two reactions: the forward (with primer TEF-264-FW) and reverse (with primer PMYR3) directions. Sequetech collecting DNA September 5, 2014 from BioCurious.
- TEF-264-FW is a custom primer synthesized through Sequetech: primer sequence recommended by DNA 2.0, suppliers of pD1214. This primer will be maintained in-house for future sequencing reactions. (Note this has changed from sequencing described August 6, 2014, as previous forward primer now anneals inside our redesigned DNA inserts.)
- PMYR3 is an in-house Sequetech primer.
- Sequencing sample hkckplus, an aliquot of August 28, 2014 midiprep, to be sequenced in two reactions: the forward (with primer TEF-264-FW) and reverse (with primer PMYR3) directions. Sequetech collecting DNA September 5, 2014 from BioCurious.
Primer Sequences PMYR3: 5' CTTCCTTTTCGGTTAGAG 3' Binds to the terminator (CYC1) and reads through in the 3'-5' direction (C-terminus of casein). TEF-264-FW: 5’ TCGATGACCTCCCATTGA 3’ Binds to TEF promoter and reads through 5'-3' (N-terminus of signal peptide).
Location of sequencing primers indicated by small purple arrows on map of pD1214: