Difference between revisions of "Sequencing Analysis using Serial Cloner 2-6-1"
Jump to navigation
Jump to search
(Created page with "= Sequencing Analysis = 1. Using Serial Cloner Freeware (2-6-1) open the forward sequencing reaction from the TEF primer. a. Search for ATG (and translate the protein by ...") |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
1. Using Serial Cloner Freeware (2-6-1) open the forward sequencing reaction from the TEF primer. | 1. Using Serial Cloner Freeware (2-6-1) open the forward sequencing reaction from the TEF primer. | ||
a. Search for ATG (and translate the protein by selection sequence from ATG onward). This provides you with a preliminary idea of whether or not your sequencing worked. You should see (at the very least) the FAKS signal peptide. Also, in the forward reaction, make sure you are using ATG on the + or 'sense' strand. | [[File:Open forward.png | 400px]] | ||
a. Search for ATG (and translate the protein by selection sequence from ATG onward). This provides you with a preliminary idea of | |||
whether or not your sequencing worked. You should see (at the very least) the FAKS signal peptide. Also, in the | |||
forward reaction, make sure you are using ATG on the + or 'sense' strand. | |||
[[File:Search for ATG.png | 400px]] | |||
b. Align the DNA to the original file using the Function>Align Two Sequences function. You will be aligning the forward sequencing | |||
reaction with the original file. I.e. Human Kappa Kex + hypothetical sequence :: Human Kappa Kex + clone 1A TEF reaction results. | |||
[[File:Function Align two seqs.png |400px]] | |||
[[File: | |||
2. Open the reverse sequence, reverse complement it using Sequence>Antiparallel function. | 2. Open the reverse sequence, reverse complement it using Sequence>Antiparallel function. | ||
a. Align this sequence to both the forward and hypothetical sequences as detailed in 1b. | a. Save the sequence as "Name Reverse Complement" | ||
b. Align this sequence to both the forward and hypothetical sequences as detailed in 1b. | |||
Latest revision as of 21:21, 16 October 2014
Sequencing Analysis
1. Using Serial Cloner Freeware (2-6-1) open the forward sequencing reaction from the TEF primer.
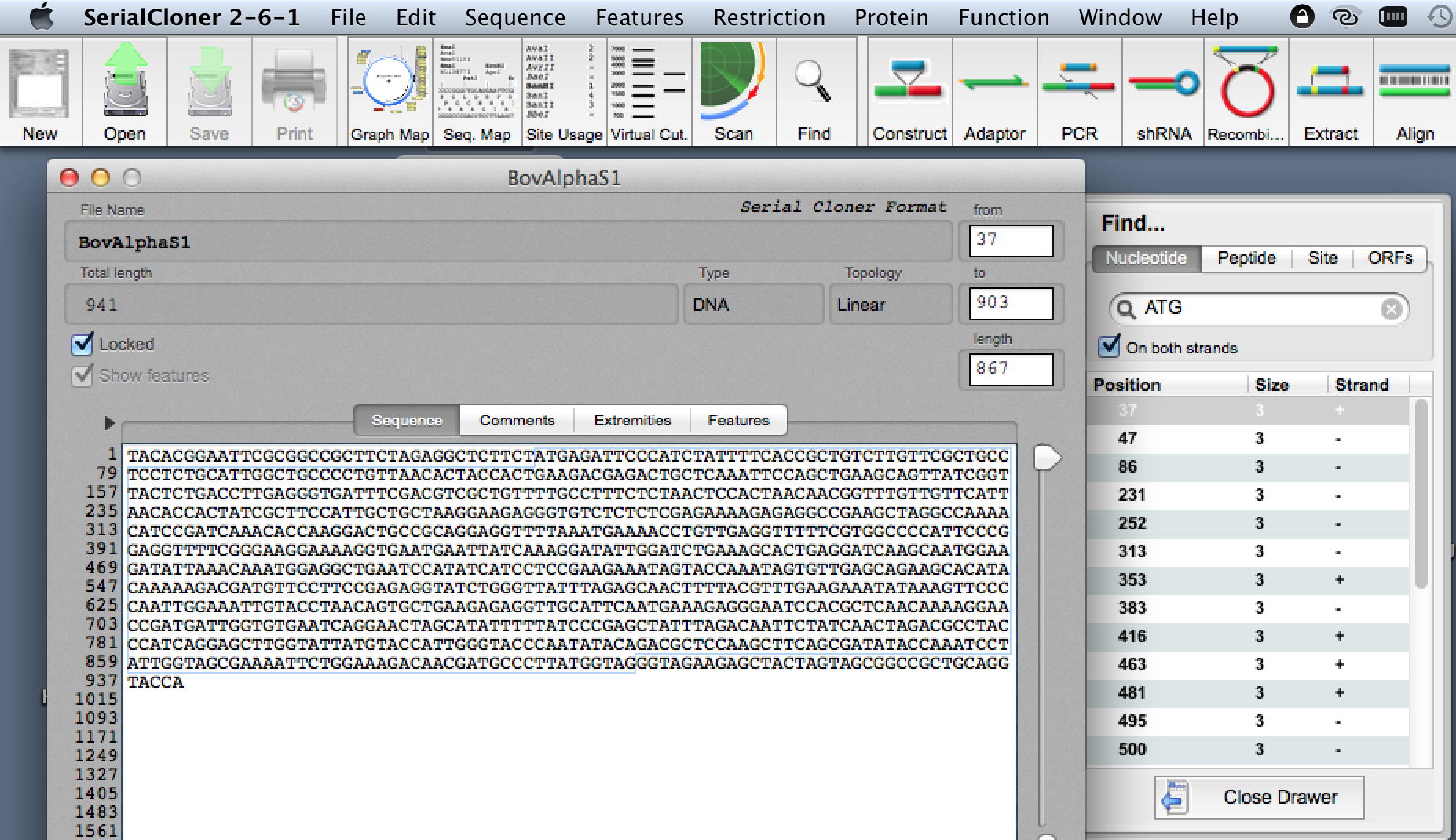
a. Search for ATG (and translate the protein by selection sequence from ATG onward). This provides you with a preliminary idea of
whether or not your sequencing worked. You should see (at the very least) the FAKS signal peptide. Also, in the
forward reaction, make sure you are using ATG on the + or 'sense' strand.
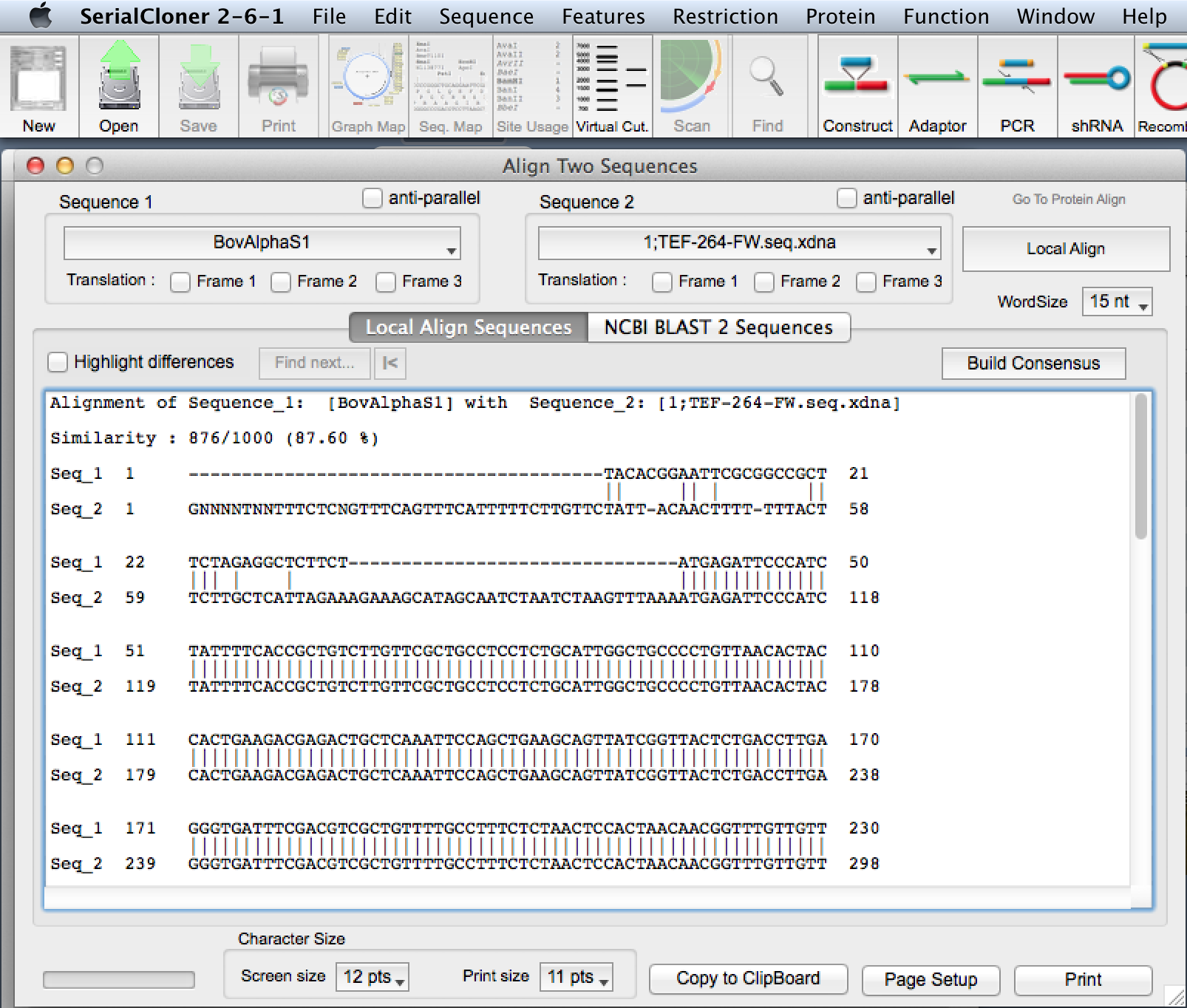
b. Align the DNA to the original file using the Function>Align Two Sequences function. You will be aligning the forward sequencing
reaction with the original file. I.e. Human Kappa Kex + hypothetical sequence :: Human Kappa Kex + clone 1A TEF reaction results.
2. Open the reverse sequence, reverse complement it using Sequence>Antiparallel function.
a. Save the sequence as "Name Reverse Complement" b. Align this sequence to both the forward and hypothetical sequences as detailed in 1b.